Which single-cell RNA sequencing data analysis tool to choose in 2024?
Some major discoveries have been made through single-cell RNA sequencing (scRNA-seq), but the process can be daunting to some scientists. Data analysis is where things get tricky; how do you process and analyze hundreds or thousands or even millions of cells without having programming skills under your belt?
Luckily, there are many scRNA-seq data analysis solutions available to you. In this article, we have compiled a list of the best tools which can help you get your single-cell analysis done in no time at all!
What is scRNA-seq?
If you aren’t familiar with scRNA-seq, it’s a method that allows you to study, you guessed it – single cells.
Cells can show very unique characteristics when examined closely, and these depend on the right genes being switched on at the right time. And scRNA-seq has made it possible to look at these unique gene signatures of individual cells.
Essentially, scRNA-seq is based on studying messenger RNA (mRNA), which reflects what proteins a cell makes and what it’s doing in specific environmental settings. This allows for the detection of differences in gene transcription between individual cells or groups of cells that would have otherwise gone overlooked. This method enables a greater understanding of the different cell populations, biological pathways, and rare cell types in a sample at a single cell resolution.
The current bottleneck of scRNA-seq research
The recent success of scRNA-seq has opened up a whole new world for scientists to explore. However, while the wet lab side of single-cell research has improved, single-cell data analysis remains a major challenge.
You need familiarity with programming languages and sufficient bioinformatics knowledge, which can take weeks or months to study!
As such there’s a growing demand for user-friendly bioinformatics solutions that can help to unveil insights locked in single-cell experiments.
scRNA-seq data analysis software: what to consider?
There are many factors to consider when choosing scRNA-seq data analysis software.
The first thing to take into consideration is the software itself – the type of application, pricing, sharing options, and whether it’s open-source.
Secondly, what can the software do for you? Is it primarily a visualization tool that’s useful only for exploring the data, or does it take you all the way from your raw data to publication-ready figures?
Below, you will find a list of factors you should keep in mind. A more extensive analysis of software features is available here!
Best scRNA-seq data analysis software
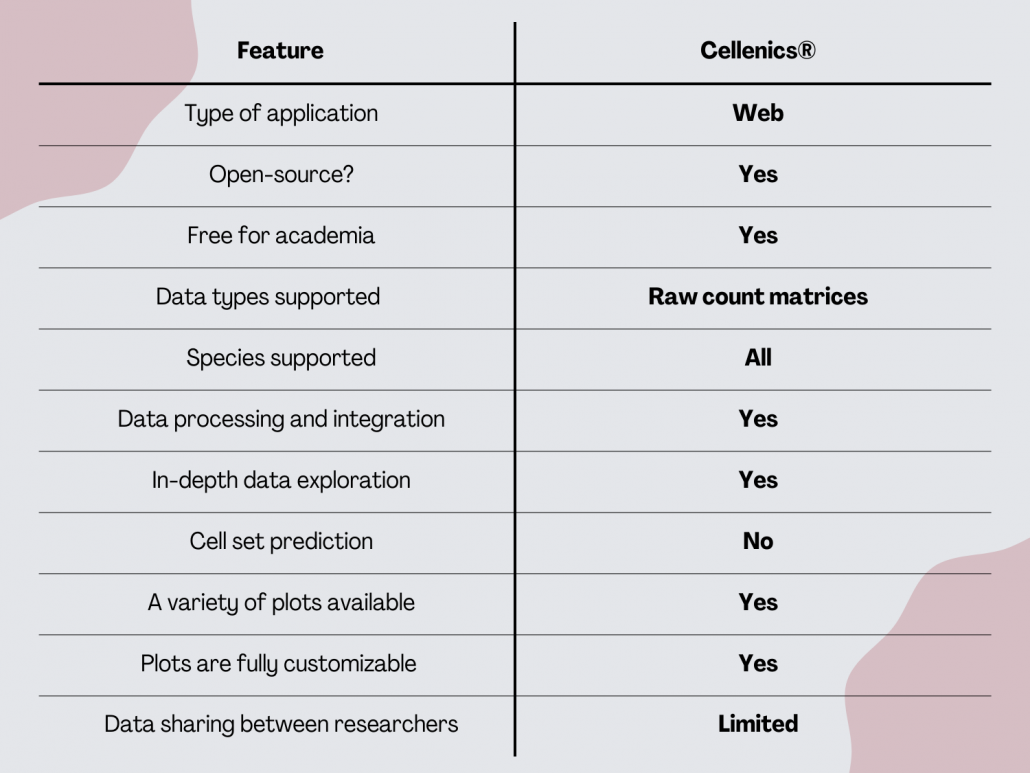
Cellenics®
Cellenics® is an open-source scRNA-seq analysis tool developed by © 2020-2022 President and Fellows of Harvard College under the scientific supervision of Prof. Peter Kharchenko and the administrative supervision of the Department of Biomedical Informatics at Harvard Medical School. Biomage is an open-source software company that provided services to Harvard Medical School for the design and development of Cellenics® and hosts a community instance of Cellenics®.
It’s a cloud-based tool that is designed specifically for biologists. Being in the cloud means scientists don’t need a powerful workstation for data analysis and storage. Cellenics® is free for academic users with datasets with up to 300,000 cells.
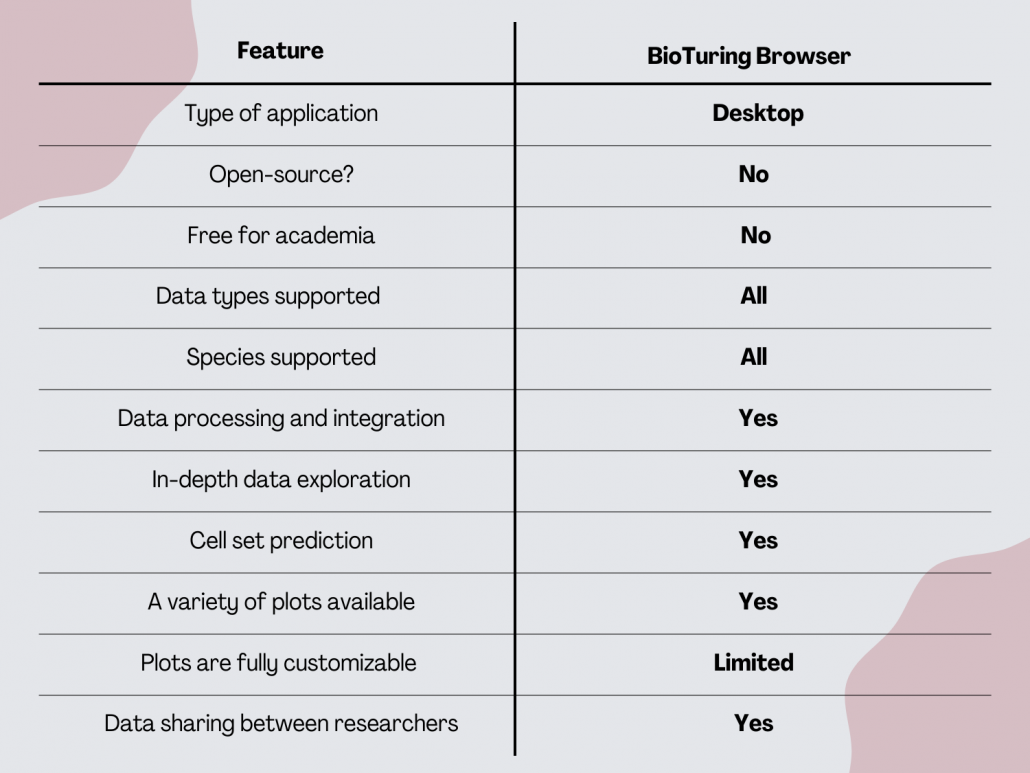
BioTuring Browser
BioTuring Browser or BBrowser offers data analysis that takes you from raw single-cell data to publication-ready figures. BBrowser is a desktop tool, where all data is stored on a local computer. This means that to analyze larger datasets, you might need to invest in a powerful workstation. BBrowser is a paid tool. Users without a license can access up to 5 public studies in the BioTuring database.
Cellxgene
Cellxgene is an open-source single-cell data analysis software that can support very large datasets with millions of cells. It’s free of charge for academic users.
It’s important to note that Cellxgene is primarily a data visualization tool, but it does offer some analysis functions. Cellxgene has a hosted data explorer and a self-hosted desktop data explorer. The downloadable self-hosted version allows the exploration of private datasets via the Python PyPI package.
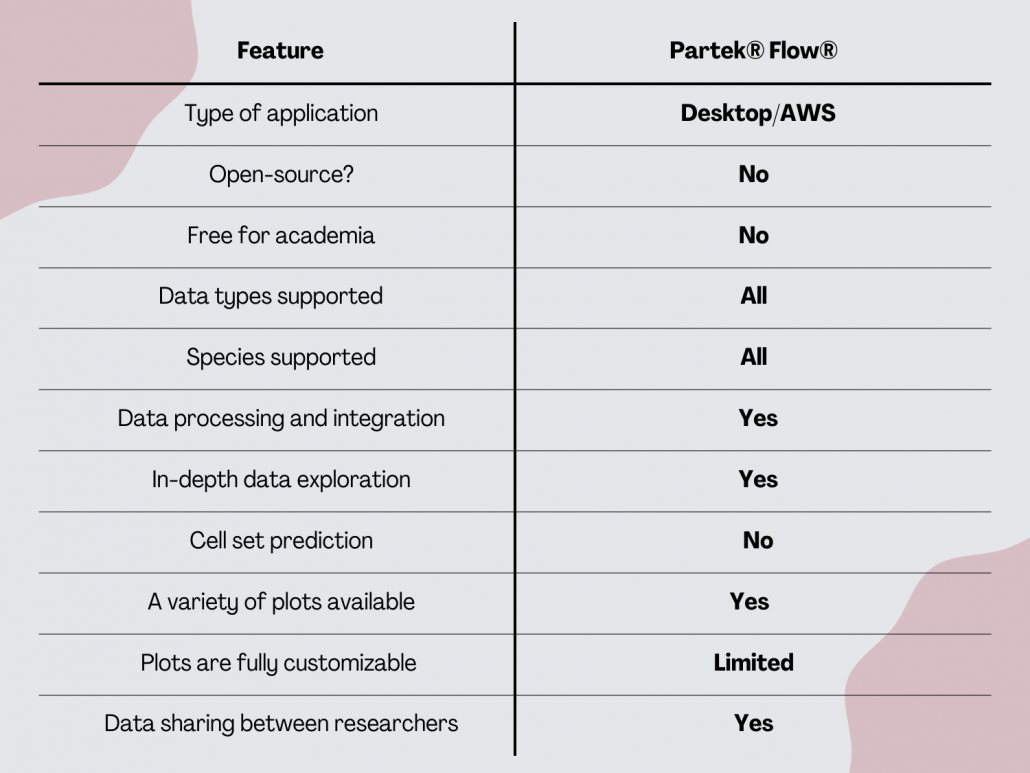
Partek® Flow®
Partek® Flow® is software for analyzing next-generation and scRNA-seq data. It is a web-based application that users can install on a desktop computer or a computer cluster. Irrespective of where the server is, the user will interact with Partek® Flow® using a web browser. Partek® Flow® is a paid software.
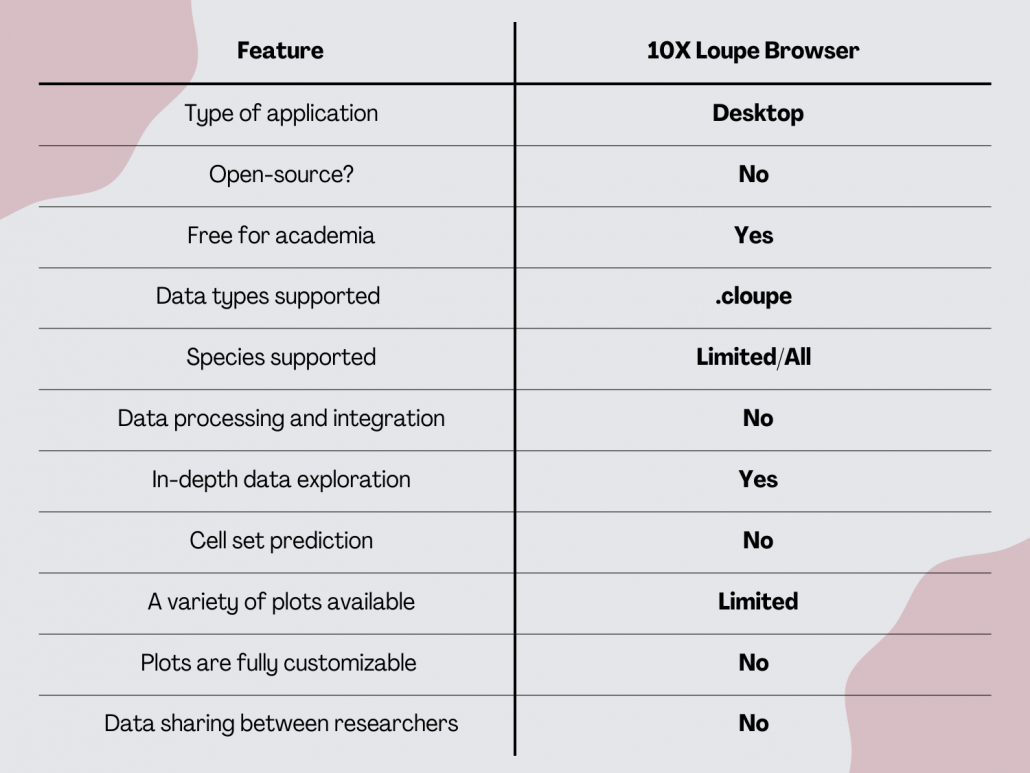
10x Loupe Browser
Loupe Browser by 10x Genomics is a desktop tool that enables anyone to analyze and visualize 10x Genomics datasets. It is worth noting that 10x Loupe Browser is not open-source software though it is available for free to all users with 10x Genomics data.
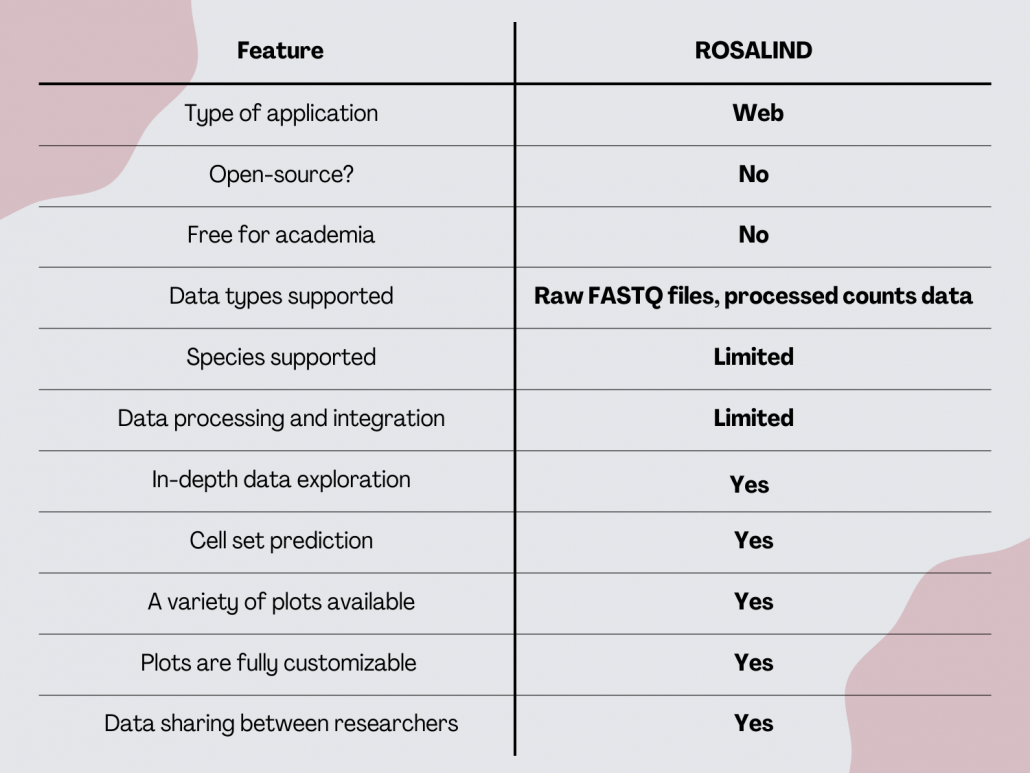
ROSALIND
ROSALIND is a cloud-based bioinformatics platform designed for life science researchers. It’s not exclusively scRNA-seq data analysis software. The software is accessible via any secure web browser. ROSALIND Spaces allows collaboration between researchers through virtual data rooms that allow to interactively explore shared experiments.
Summary
scRNA-seq analysis can seem intimidating, but with the right tool, anyone without any coding experience or access to a bioinformatician can work with single-cell data. You will be able to gain biological insight in as little as a few hours from your datasets, and take the next step in advancing your research project.
The decision on which tool to use for analyzing scRNA-seq data is not universal, as each has its own advantages and disadvantages. The information presented here should help you to make a more informed choice and more confident in choosing one to analyze your single-cell scRNA-seq data.
A more in-depth article that includes a breakdown of the factors and each tool in more detail is available here: https://www.biomage.net/blog/best-single-cell-rna-sequencing-data-analysis-tools-in-2022.
Conflict of interest disclaimer.
Biomage hosts a community instance of Cellenics®, an open-source, cloud-based analytics tool for single-cell RNA-sequencing data. Cellenics® has been included in our list of the best scRNA-seq data analysis tools.